Serialization and IO¶
xray supports direct serialization and IO to several file formats. For more options, consider exporting your objects to pandas (see the preceding section) and using its broad range of IO tools.
Pickle¶
The simplest way to serialize an xray object is to use Python’s built-in pickle module:
In [1]: import cPickle as pickle
In [2]: ds = xray.Dataset({'foo': (('x', 'y'), np.random.rand(4, 5))},
...: coords={'x': [10, 20, 30, 40],
...: 'y': pd.date_range('2000-01-01', periods=5),
...: 'z': ('x', list('abcd'))})
...:
# use the highest protocol (-1) because it is way faster than the default
# text based pickle format
In [3]: pkl = pickle.dumps(ds, protocol=-1)
In [4]: pickle.loads(pkl)
Out[4]:
<xray.Dataset>
Dimensions: (x: 4, y: 5)
Coordinates:
* y (y) datetime64[ns] 2000-01-01 2000-01-02 2000-01-03 2000-01-04 ...
* x (x) int64 10 20 30 40
z (x) |S1 'a' 'b' 'c' 'd'
Data variables:
foo (x, y) float64 0.127 0.9667 0.2605 0.8972 0.3767 0.3362 0.4514 ...
Pickle support is important because it doesn’t require any external libraries and lets you use xray objects with Python modules like multiprocessing. However, there are two important caveats:
- To simplify serialization, xray’s support for pickle currently loads all array values into memory before dumping an object. This means it is not suitable for serializing datasets too big to load into memory (e.g., from netCDF or OPeNDAP).
- Pickle will only work as long as the internal data structure of xray objects remains unchanged. Because the internal design of xray is still being refined, we make no guarantees (at this point) that objects pickled with this version of xray will work in future versions.
netCDF¶
Currently, the only disk based serialization format that xray directly supports is netCDF. netCDF is a file format for fully self-described datasets that is widely used in the geosciences and supported on almost all platforms. We use netCDF because xray was based on the netCDF data model, so netCDF files on disk directly correspond to Dataset objects. Recent versions netCDF are based on the even more widely used HDF5 file-format.
Reading and writing netCDF files with xray requires the netCDF4-Python library or scipy to be installed.
We can save a Dataset to disk using the Dataset.to_netcdf method:
In [5]: ds.to_netcdf('saved_on_disk.nc')
By default, the file is saved as netCDF4 (assuming netCDF4-Python is installed). You can control the format and engine used to write the file with the format and engine arguments.
We can load netCDF files to create a new Dataset using open_dataset():
In [6]: ds_disk = xray.open_dataset('saved_on_disk.nc')
In [7]: ds_disk
Out[7]:
<xray.Dataset>
Dimensions: (x: 4, y: 5)
Coordinates:
* y (y) datetime64[ns] 2000-01-01 2000-01-02 2000-01-03 2000-01-04 ...
* x (x) >i4 10 20 30 40
z (x) |S1 'a' 'b' 'c' 'd'
Data variables:
foo (x, y) >f4 0.12697 0.966718 0.260476 0.897237 0.37675 0.336222 ...
A dataset can also be loaded or written to a specific group within a netCDF file. To load from a group, pass a group keyword argument to the open_dataset function. The group can be specified as a path-like string, e.g., to access subgroup ‘bar’ within group ‘foo’ pass ‘/foo/bar’ as the group argument. When writing multiple groups in one file, pass mode='a' to to_netcdf to ensure that each call does not delete the file.
Data is loaded lazily from netCDF files. You can manipulate, slice and subset Dataset and DataArray objects, and no array values are loaded into memory until you try to perform some sort of actual computation. For an example of how these lazy arrays work, see the OPeNDAP section below.
It is important to note that when you modify values of a Dataset, even one linked to files on disk, only the in-memory copy you are manipulating in xray is modified: the original file on disk is never touched.
Tip
xray’s lazy loading of remote or on-disk datasets is often but not always desirable. Before performing computationally intense operations, it is usually a good idea to load a dataset entirely into memory by invoking the load() method.
Datasets have a close() method to close the associated netCDF file. However, it’s often cleaner to use a with statement:
# this automatically closes the dataset after use
In [8]: with xray.open_dataset('saved_on_disk.nc') as ds:
...: print(ds.keys())
...:
['y', 'x', 'z', 'foo']
Note
Although xray provides reasonable support for incremental reads of files on disk, it does not yet support incremental writes, which is important for dealing with datasets that do not fit into memory. This is a significant shortcoming that we hope to resolve (GH199) by adding the ability to create Dataset objects directly linked to a netCDF file on disk.
NetCDF files follow some conventions for encoding datetime arrays (as numbers with a “units” attribute) and for packing and unpacking data (as described by the “scale_factor” and “_FillValue” attributes). If the argument decode_cf=True (default) is given to open_dataset, xray will attempt to automatically decode the values in the netCDF objects according to CF conventions. Sometimes this will fail, for example, if a variable has an invalid “units” or “calendar” attribute. For these cases, you can turn this decoding off manually.
You can view this encoding information and control the details of how xray serializes objects, by viewing and manipulating the DataArray.encoding attribute:
In [9]: ds_disk['y'].encoding
Out[9]:
{'calendar': u'proleptic_gregorian',
'chunksizes': None,
'complevel': 0,
'contiguous': True,
'dtype': dtype('float64'),
'fletcher32': False,
'least_significant_digit': None,
'shuffle': False,
'source': 'saved_on_disk.nc',
'units': u'days since 2000-01-01 00:00:00',
'zlib': False}
OPeNDAP¶
xray includes support for OPeNDAP (via the netCDF4 library or Pydap), which lets us access large datasets over HTTP.
For example, we can open a connection to GBs of weather data produced by the PRISM project, and hosted by IRI at Columbia:
In [10]: remote_data = xray.open_dataset(
....: 'http://iridl.ldeo.columbia.edu/SOURCES/.OSU/.PRISM/.monthly/dods',
....: decode_times=False)
....:
In [11]: remote_data
Out[11]:
<xray.Dataset>
Dimensions: (T: 1422, X: 1405, Y: 621)
Coordinates:
* X (X) float32 -125.0 -124.958 -124.917 -124.875 -124.833 -124.792 -124.75 ...
* T (T) float32 -779.5 -778.5 -777.5 -776.5 -775.5 -774.5 -773.5 -772.5 -771.5 ...
* Y (Y) float32 49.9167 49.875 49.8333 49.7917 49.75 49.7083 49.6667 49.625 ...
Data variables:
ppt (T, Y, X) float64 ...
tdmean (T, Y, X) float64 ...
tmax (T, Y, X) float64 ...
tmin (T, Y, X) float64 ...
Attributes:
Conventions: IRIDL
expires: 1375315200
Note
Like many real-world datasets, this dataset does not entirely follow CF conventions. Unexpected formats will usually cause xray’s automatic decoding to fail. The way to work around this is to either set decode_cf=False in open_dataset to turn off all use of CF conventions, or by only disabling the troublesome parser. In this case, we set decode_times=False because the time axis here provides the calendar attribute in a format that xray does not expect (the integer 360 instead of a string like '360_day').
We can select and slice this data any number of times, and nothing is loaded over the network until we look at particular values:
In [12]: tmax = remote_data['tmax'][:500, ::3, ::3]
In [13]: tmax
Out[13]:
<xray.DataArray 'tmax' (T: 500, Y: 207, X: 469)>
[48541500 values with dtype=float64]
Coordinates:
* Y (Y) float32 49.9167 49.7917 49.6667 49.5417 49.4167 49.2917 49.1667 ...
* X (X) float32 -125.0 -124.875 -124.75 -124.625 -124.5 -124.375 -124.25 ...
* T (T) float32 -779.5 -778.5 -777.5 -776.5 -775.5 -774.5 -773.5 -772.5 -771.5 ...
Attributes:
pointwidth: 120
standard_name: air_temperature
units: Celsius_scale
expires: 1375315200
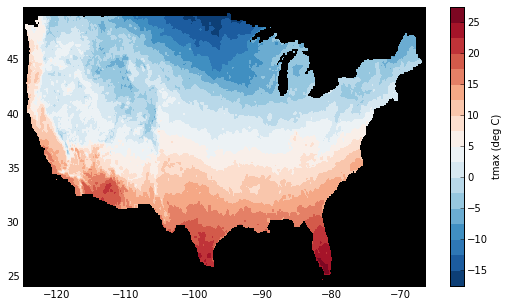
Finally, let’s plot a small subset with matplotlib:
In [14]: tmax_ss = tmax[0]
In [15]: import matplotlib.pyplot as plt
In [16]: plt.figure(figsize=(9, 5))
In [17]: plt.gca().patch.set_color('0')
In [18]: plt.contourf(tmax_ss['X'], tmax_ss['Y'], tmax_ss.values, 20,
....: cmap='RdBu_r')
....:
In [19]: plt.colorbar(label='tmax (deg C)')
Note
We do hope to eventually add plotting methods to xray to make this easier (GH185).
Combining multiple files¶
NetCDF files are often encountered in collections, e.g., with different files corresponding to different model runs. xray can straightforwardly combine such files into a single Dataset by making use of concat().
Note
Version 0.5 includes experimental support for manipulating datasets that don’t fit into memory with dask. If you have dask installed, you can open multiple files simultaneously using open_mfdataset():
xray.open_mfdataset('my/files/*.nc')
This function automatically concatenates and merges into a single xray datasets. For more details, see Reading and writing data.
For example, here’s how we could approximate MFDataset from the netCDF4 library:
from glob import glob
import xray
def read_netcdfs(files, dim):
# glob expands paths with * to a list of files, like the unix shell
paths = sorted(glob(files))
datasets = [xray.open_dataset(p) for p in paths]
combined = xray.concat(dataset, dim)
return combined
read_netcdfs('/all/my/files/*.nc', dim='time')
This function will work in many cases, but it’s not very robust. First, it never closes files, which means it will fail one you need to load more than a few thousands file. Second, it assumes that you want all the data from each file and that it can all fit into memory. In many situations, you only need a small subset or an aggregated summary of the data from each file.
Here’s a slightly more sophisticated example of how to remedy these deficiencies:
def read_netcdfs(files, dim, transform_func=None):
def process_one_path(path):
# use a context manager, to ensure the file gets closed after use
with xray.open_dataset(path) as ds:
# transform_func should do some sort of selection or
# aggregation
if transform_func is not None:
ds = transform_func(ds)
# load all data from the transformed dataset, to ensure we can
# use it after closing each original file
ds.load()
return ds
paths = sorted(glob(files))
datasets = [process_one_path(p) for p in paths]
xray.concat(dataset, dim)
# here we suppose we only care about the combined mean of each file;
# you might also use indexing operations like .sel to subset datasets
read_netcdfs('/all/my/files/*.nc', dim='time',
transform_func=lambda ds: ds.mean())
This pattern works well and is very robust. We’ve used similar code to process tens of thousands of files constituting 100s of GB of data.